Xarray Tips and Tricks
Contents
Xarray Tips and Tricks#
Build a multi-file dataset from an OpenDAP server#
One thing we love about xarray is the open_mfdataset function, which combines many netCDF files into a single xarray Dataset.
But what if the files are stored on a remote server and accessed over OpenDAP. An example can be found in NOAA’s NCEP Reanalysis catalog.
https://www.esrl.noaa.gov/psd/thredds/catalog/Datasets/ncep.reanalysis/surface/catalog.html
The dataset is split into different files for each variable and year. For example, a single file for surface air temperature looks like:
http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1948.nc
We can’t just call
open_mfdataset("http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.*.nc")
Because wildcard expansion doesn’t work with OpenDAP endpoints. The solution is to manually create a list of files to open.
base_url = 'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995'
files = [f'{base_url}.{year}.nc' for year in range(1948, 2019)]
files
['http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1948.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1949.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1950.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1951.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1952.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1953.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1954.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1955.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1956.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1957.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1958.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1959.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1960.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1961.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1962.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1963.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1964.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1965.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1966.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1967.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1968.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1969.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1970.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1971.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1972.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1973.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1974.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1975.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1976.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1977.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1978.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1979.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1980.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1981.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1982.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1983.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1984.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1985.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1986.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1987.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1988.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1989.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1990.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1991.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1992.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1993.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1994.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1995.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1996.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1997.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1998.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.1999.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2000.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2001.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2002.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2003.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2004.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2005.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2006.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2007.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2008.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2009.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2010.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2011.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2012.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2013.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2014.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2015.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2016.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2017.nc',
'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/ncep.reanalysis/surface/air.sig995.2018.nc']
import xarray as xr
%matplotlib inline
ds = xr.open_mfdataset(files)
ds
<xarray.Dataset>
Dimensions: (lat: 73, lon: 144, time: 103596)
Coordinates:
* lon (lon) float32 0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 ...
* lat (lat) float32 90.0 87.5 85.0 82.5 80.0 77.5 75.0 72.5 70.0 67.5 ...
* time (time) datetime64[ns] 1948-01-01 1948-01-01T06:00:00 ...
Data variables:
air (time, lat, lon) float32 dask.array<shape=(103596, 73, 144), chunksize=(1464, 73, 144)>
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\...
platform: Model
history: created 99/05/11 by Hoop (netCDF2.3)\nCo...
References: http://www.esrl.noaa.gov/psd/data/gridde...
dataset_title: NCEP-NCAR Reanalysis 1
DODS_EXTRA.Unlimited_Dimension: time
We have now opened the entire ensemble of files on the remote server as a single xarray Dataset!
Perform Correlation Analysis#
Many people want to look at correlations (usually in time) in their final projects. Unfortunately, this is not currently part of xarray (but it will be added soon! – see open issue).
In the meantime, the following functions works pretty well.
def covariance(x, y, dims=None):
return xr.dot(x - x.mean(dims), y - y.mean(dims), dims=dims) / x.count(dims)
def corrrelation(x, y, dims=None):
return covariance(x, y, dims) / (x.std(dims) * y.std(dims))
ds = xr.open_dataset('NOAA_NCDC_ERSST_v3b_SST.nc')
ds
<xarray.Dataset>
Dimensions: (lat: 89, lon: 180, time: 684)
Coordinates:
* lat (lat) float32 -88.0 -86.0 -84.0 -82.0 -80.0 -78.0 -76.0 -74.0 ...
* lon (lon) float32 0.0 2.0 4.0 6.0 8.0 10.0 12.0 14.0 16.0 18.0 20.0 ...
* time (time) datetime64[ns] 1960-01-15 1960-02-15 1960-03-15 ...
Data variables:
sst (time, lat, lon) float32 ...
Attributes:
Conventions: IRIDL
source: https://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NCDC/.ERSST/...
history: extracted and cleaned by Ryan Abernathey for Research Compu...
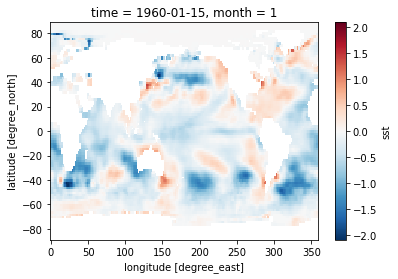
sst = ds.sst
sst_clim = sst.groupby('time.month').mean(dim='time')
sst_anom = sst.groupby('time.month') - sst_clim
%matplotlib inline
sst_anom[0].plot()
<matplotlib.collections.QuadMesh at 0x7fc6018af5f8>
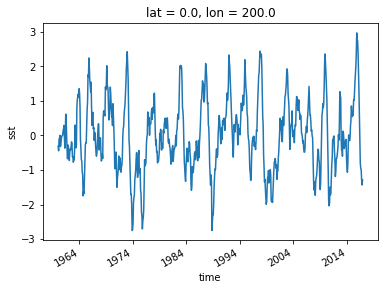
sst_ref = sst_anom.sel(lon=200, lat=0, method='nearest')
sst_ref.plot()
[<matplotlib.lines.Line2D at 0x7fc6017de7f0>]
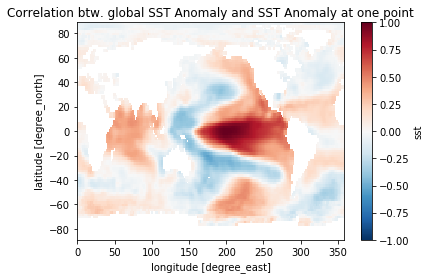
sst_cor = corrrelation(sst_anom, sst_ref, dims='time')
pc = sst_cor.plot()
pc.axes.set_title('Correlation btw. global SST Anomaly and SST Anomaly at one point')
Text(0.5,1,'Correlation btw. global SST Anomaly and SST Anomaly at one point')
Fix poorly encoded time coordinates#
Many datasets, especially from INGRID, are encoded with “months since” as the time unit. Trying to open these in xarray currently gives an error.
import cftime
cftime.__version__
'1.0.3'
import xarray as xr
url = 'http://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NCEP/.CPC/.CAMS_OPI/.v0208/.mean/.prcp/dods'
ds = xr.open_dataset(url)
---------------------------------------------------------------------------
OutOfBoundsDatetime Traceback (most recent call last)
/opt/conda/lib/python3.6/site-packages/xarray/coding/times.py in decode_cf_datetime(num_dates, units, calendar, enable_cftimeindex)
172 if calendar not in _STANDARD_CALENDARS:
--> 173 raise OutOfBoundsDatetime
174
OutOfBoundsDatetime:
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
/opt/conda/lib/python3.6/site-packages/xarray/coding/times.py in _decode_cf_datetime_dtype(data, units, calendar, enable_cftimeindex)
131 result = decode_cf_datetime(example_value, units, calendar,
--> 132 enable_cftimeindex)
133 except Exception:
/opt/conda/lib/python3.6/site-packages/xarray/coding/times.py in decode_cf_datetime(num_dates, units, calendar, enable_cftimeindex)
200 flat_num_dates.astype(np.float), units, calendar,
--> 201 enable_cftimeindex)
202
/opt/conda/lib/python3.6/site-packages/xarray/coding/times.py in _decode_datetime_with_cftime(num_dates, units, calendar, enable_cftimeindex)
94 else:
---> 95 dates = np.asarray(cftime.num2date(num_dates, units, calendar))
96
cftime/_cftime.pyx in cftime._cftime.num2date()
cftime/_cftime.pyx in cftime._cftime.utime.__init__()
ValueError: calendar must be one of ['standard', 'gregorian', 'proleptic_gregorian', 'noleap', 'julian', 'all_leap', '365_day', '366_day', '360_day'], got '360'
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
<ipython-input-9-95c7bea581ab> in <module>()
2
3 url = 'http://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NCEP/.CPC/.CAMS_OPI/.v0208/.mean/.prcp/dods'
----> 4 ds = xr.open_dataset(url)
/opt/conda/lib/python3.6/site-packages/xarray/backends/api.py in open_dataset(filename_or_obj, group, decode_cf, mask_and_scale, decode_times, autoclose, concat_characters, decode_coords, engine, chunks, lock, cache, drop_variables, backend_kwargs)
344 lock = _default_lock(filename_or_obj, engine)
345 with close_on_error(store):
--> 346 return maybe_decode_store(store, lock)
347 else:
348 if engine is not None and engine != 'scipy':
/opt/conda/lib/python3.6/site-packages/xarray/backends/api.py in maybe_decode_store(store, lock)
256 store, mask_and_scale=mask_and_scale, decode_times=decode_times,
257 concat_characters=concat_characters, decode_coords=decode_coords,
--> 258 drop_variables=drop_variables)
259
260 _protect_dataset_variables_inplace(ds, cache)
/opt/conda/lib/python3.6/site-packages/xarray/conventions.py in decode_cf(obj, concat_characters, mask_and_scale, decode_times, decode_coords, drop_variables)
427 vars, attrs, coord_names = decode_cf_variables(
428 vars, attrs, concat_characters, mask_and_scale, decode_times,
--> 429 decode_coords, drop_variables=drop_variables)
430 ds = Dataset(vars, attrs=attrs)
431 ds = ds.set_coords(coord_names.union(extra_coords).intersection(vars))
/opt/conda/lib/python3.6/site-packages/xarray/conventions.py in decode_cf_variables(variables, attributes, concat_characters, mask_and_scale, decode_times, decode_coords, drop_variables)
360 k, v, concat_characters=concat_characters,
361 mask_and_scale=mask_and_scale, decode_times=decode_times,
--> 362 stack_char_dim=stack_char_dim)
363 if decode_coords:
364 var_attrs = new_vars[k].attrs
/opt/conda/lib/python3.6/site-packages/xarray/conventions.py in decode_cf_variable(name, var, concat_characters, mask_and_scale, decode_times, decode_endianness, stack_char_dim)
298 for coder in [times.CFTimedeltaCoder(),
299 times.CFDatetimeCoder()]:
--> 300 var = coder.decode(var, name=name)
301
302 dimensions, data, attributes, encoding = (
/opt/conda/lib/python3.6/site-packages/xarray/coding/times.py in decode(self, variable, name)
402 calendar = pop_to(attrs, encoding, 'calendar')
403 dtype = _decode_cf_datetime_dtype(
--> 404 data, units, calendar, enable_cftimeindex)
405 transform = partial(
406 decode_cf_datetime, units=units, calendar=calendar,
/opt/conda/lib/python3.6/site-packages/xarray/coding/times.py in _decode_cf_datetime_dtype(data, units, calendar, enable_cftimeindex)
139 if not PY3:
140 msg += ' Full traceback:\n' + traceback.format_exc()
--> 141 raise ValueError(msg)
142 else:
143 dtype = getattr(result, 'dtype', np.dtype('object'))
ValueError: unable to decode time units 'months since 1960-01-01' with calendar '360'. Try opening your dataset with decode_times=False.
One way to avoid this is to simply not decode the times:
ds = xr.open_dataset(url, decode_times=False)
ds
<xarray.Dataset>
Dimensions: (T: 478, X: 144, Y: 72)
Coordinates:
* X (X) float32 1.25 3.75 6.25 8.75 11.25 13.75 16.25 18.75 21.25 ...
* T (T) float32 228.5 229.5 230.5 231.5 232.5 233.5 234.5 235.5 ...
* Y (Y) float32 -88.75 -86.25 -83.75 -81.25 -78.75 -76.25 -73.75 ...
Data variables:
prcp (T, Y, X) float32 ...
Attributes:
Conventions: IRIDL
However, this is not satisfying, because now we can’t use xarray’s time aware features (like resample).
A better option has recently become possible. First we need the latest development version of the cftime package.
def fix_calendar(ds, timevar='T'):
if ds[timevar].attrs['calendar'] == '360':
ds[timevar].attrs['calendar'] = '360_day'
return ds
ds = fix_calendar(ds)
ds = xr.decode_cf(ds)
ds
<xarray.Dataset>
Dimensions: (T: 478, X: 144, Y: 72)
Coordinates:
* X (X) float32 1.25 3.75 6.25 8.75 11.25 13.75 16.25 18.75 21.25 ...
* T (T) datetime64[ns] 1979-01-16 1979-02-16 1979-03-16 1979-04-16 ...
* Y (Y) float32 -88.75 -86.25 -83.75 -81.25 -78.75 -76.25 -73.75 ...
Data variables:
prcp (T, Y, X) float32 ...
Attributes:
Conventions: IRIDL